Structural analysis of alloantibody-HLA interactions and of HLA B-cell epitopes
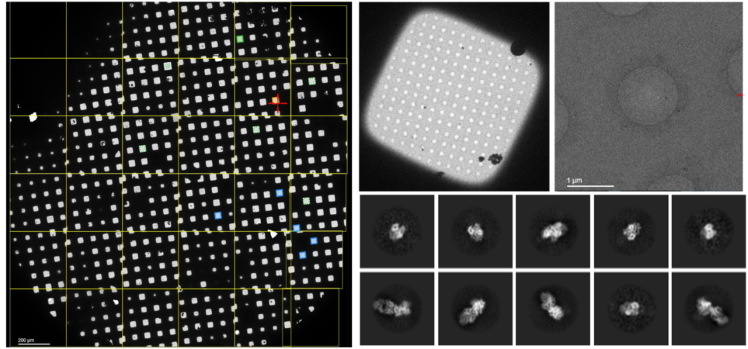
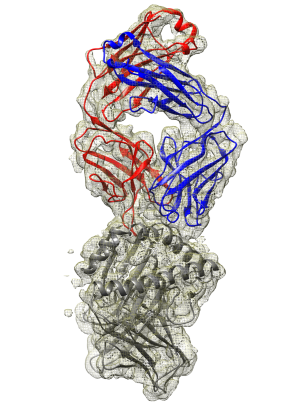
We are interested in understanding the structure of immunogenic HLA B-cell epitopes that drive humoral alloimmune responses in transplant recipients and study the molecular basis of alloantibody-HLA interactions. We employ computational techniques (e.g. HLA and antibody structure modelling, molecular dynamics simulations) and cryo-Electron Microscopy (collaboration with the European Molecular Biology Laboratory in Hinxton UK and Grenoble France) in combination with in vitro cellular assays, HLA-specific B-cell isolation and recombinant HLA monoclonal antibody generation (in collaboration with the Transplant Immunology Group in Leiden, Netherlands), and direct quantification of alloantibody-HLA interactions.
We have created an efficient pipeline for structural determination of multiple antibody-HLA complexes using cryo-EM and aim to generate a library of immunogenic HLA B-cell epitopes to define HLA compatibility at the epitope level.
Key References
Mallon DH, Bradley JA, Winn PJ, Taylor CJ, Kosmoliaptsis V. Three-dimensional structural modelling and calculation of electrostatic potentials of HLA Bw4 and Bw6 epitopes to explain the molecular basis for alloantibody binding: toward predicting HLA antigenicity and immunogenicity. Transplantation 2015; 99: 385
Goyette P, Boucher G, Mallon D et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015; 47: 172
Kosmoliaptsis V, Dafforn TR, Chaudhry AN, Halsall DJ, Bradley JA, Taylor CJ. High-resolution, three-dimensional modeling of human leukocyte antigen class I structure and surface electrostatic potential reveals the molecular basis for alloantibody binding epitopes. Hum Immunol. 2011; 72: 1049